We study soil fungi that cause disease when fungal cells are inhaled by mammalian hosts, including people.
Areas of interest:
Microbial Signal Transduction
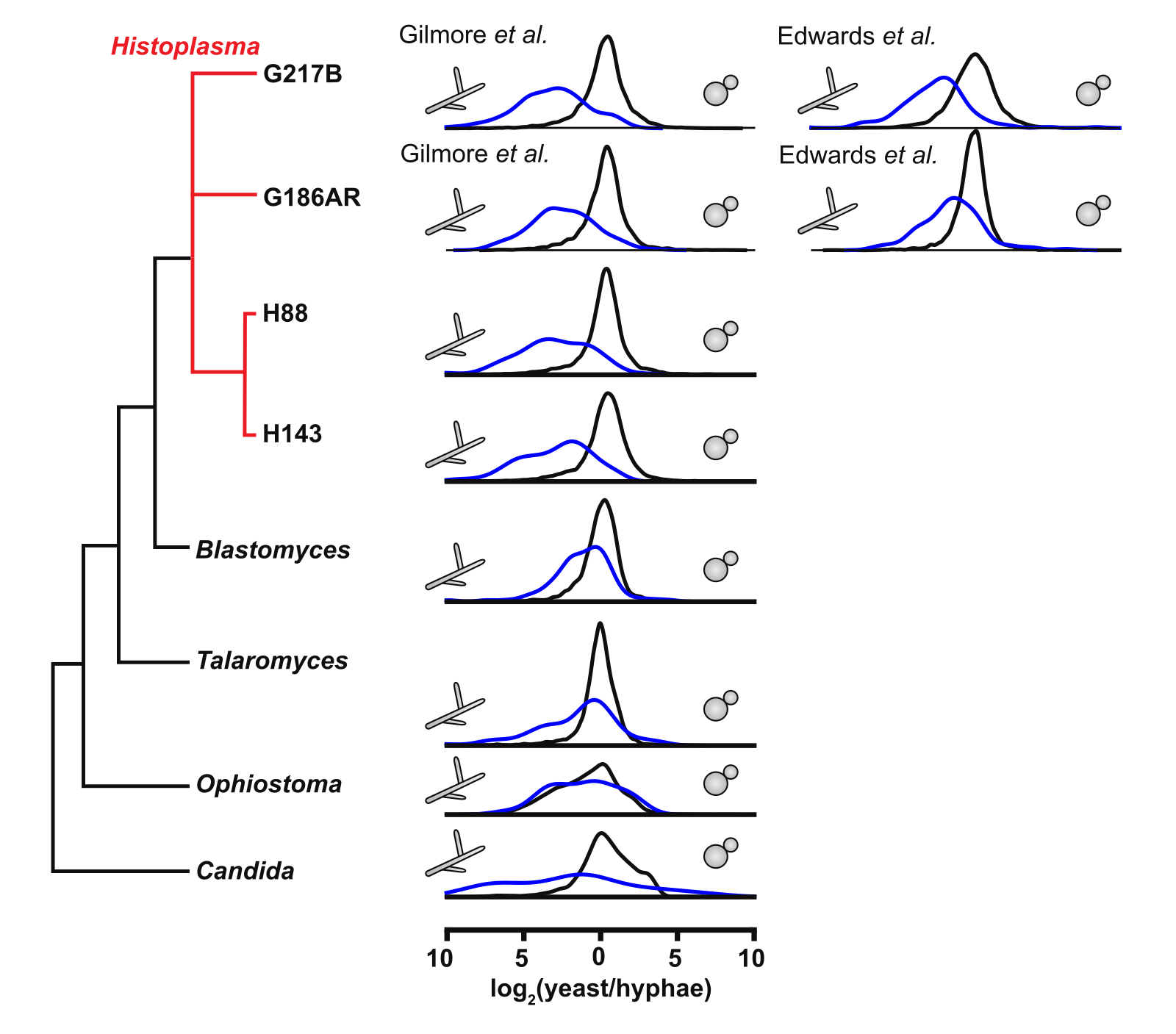
How do microbial cells sense temperature? Thermally dimorphic fungi such as Histoplasma and Coccidioides species are primary pathogens that cause disease in both healthy and immunocompromised hosts. These pathogenic fungi undergo drastic transitions in cell shape and gene expression when exposed to different temperatures, and host temperature is the key signal that triggers a developmental pathway resulting in cell shape changes and virulence gene expression required to cause disease. We use genetics and functional genomics to elucidate molecules that sense and respond to temperature.
Comparative Genomics
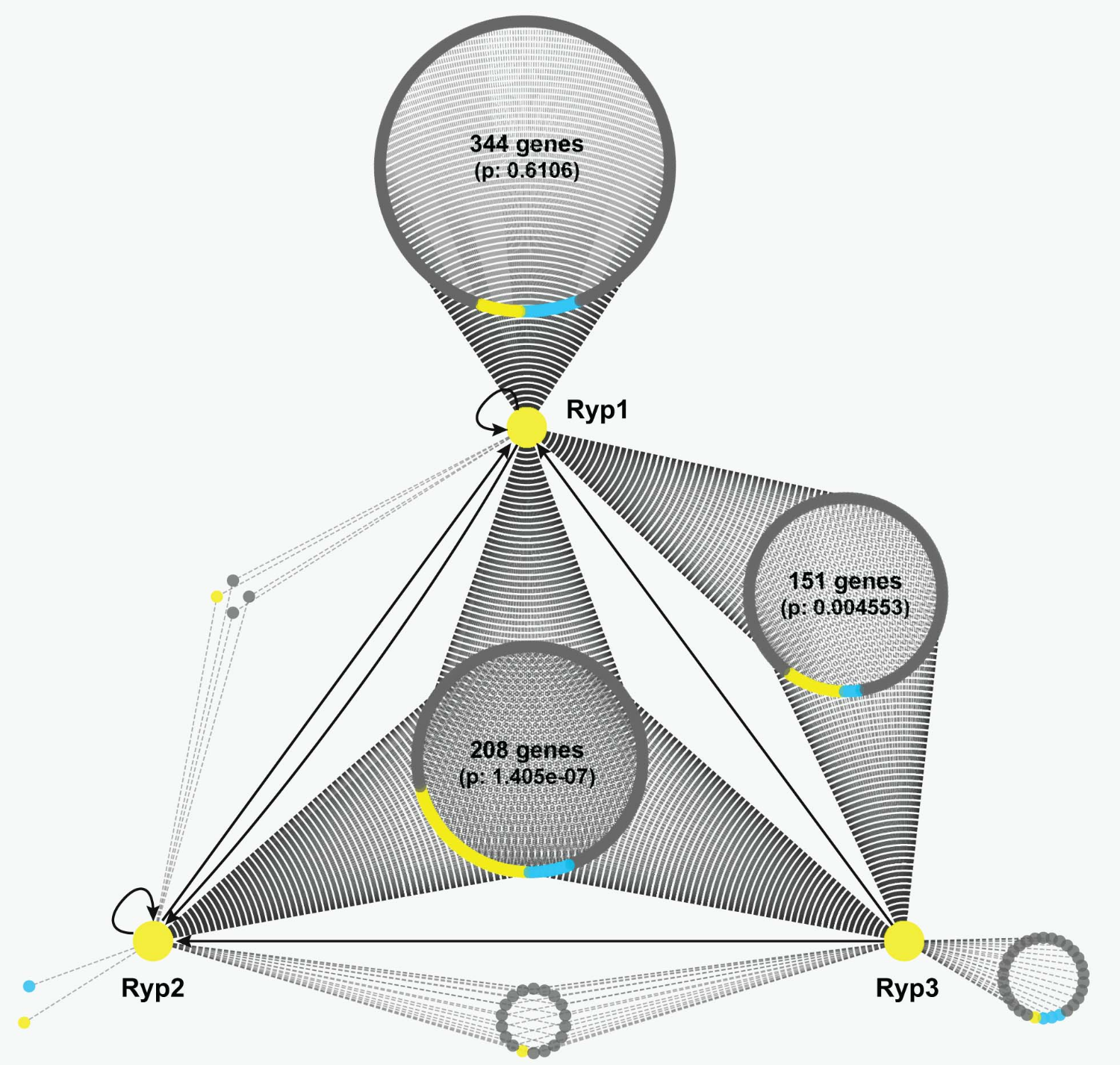
How do fungi specify cell fate? Fungal developmental programs require multiple regulatory proteins that have been rewired across the fungal kingdom to respond to different inputs. As experts in transcriptomics, we study how transcriptional networks and other regulatory modules function across fungal species to trigger key cell fate decisions.
Innate Immunity
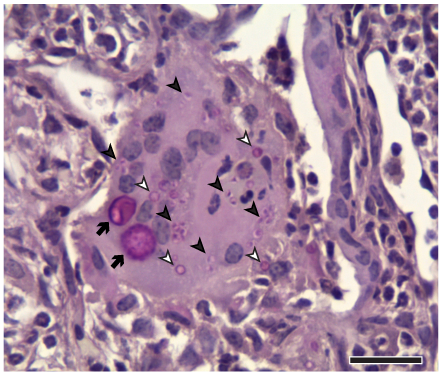
How do fungal pathogens counter the innate immune response? Histoplasma and Coccidioides species can subvert the innate immune response to replicate and cause disease. We study the ability of Histoplasma to evade anti-microbial defenses and colonize macrophages.
Cell Biology
How do intracellular fungi subvert the normal cell biology of the macrophage? Histoplasma replicates within the macrophage phagosome, and unknown mechanisms are used to recruit host membrane around each microbial cell as it divides. We want to understand how Histoplasma manipulates the fundamental biology of the host cell.
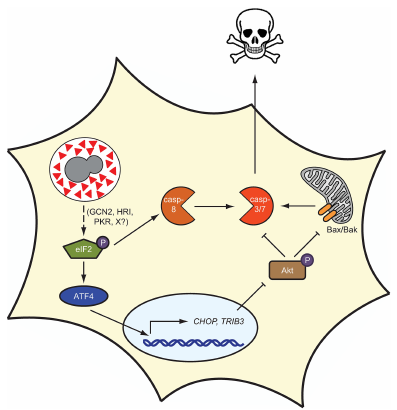
Mammalian Cell Death
How do intracellular fungi exit their host cell? We have shown that the ability of Histoplasma to trigger host cell lysis is dependent on secreted effectors from the fungus. We study how these effectors trigger cellular stress and host cell death.